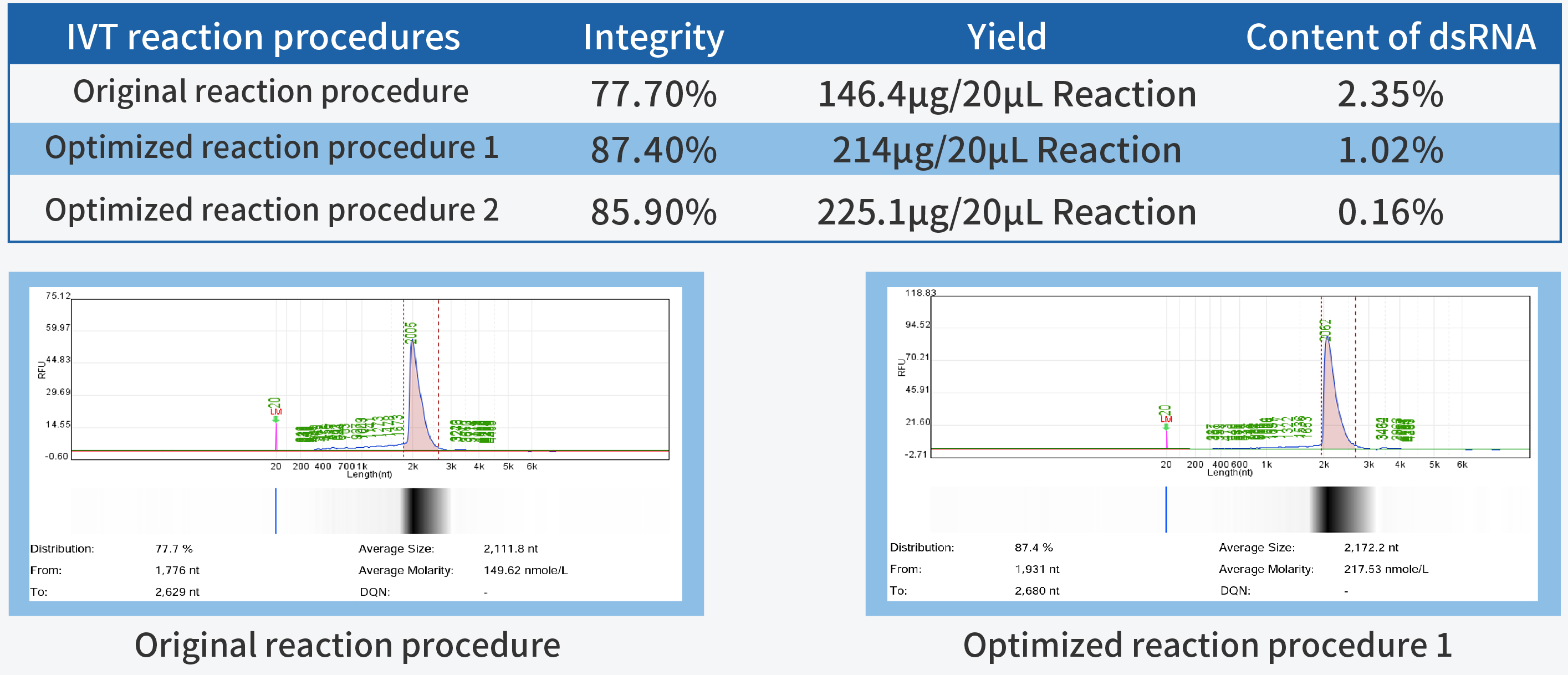
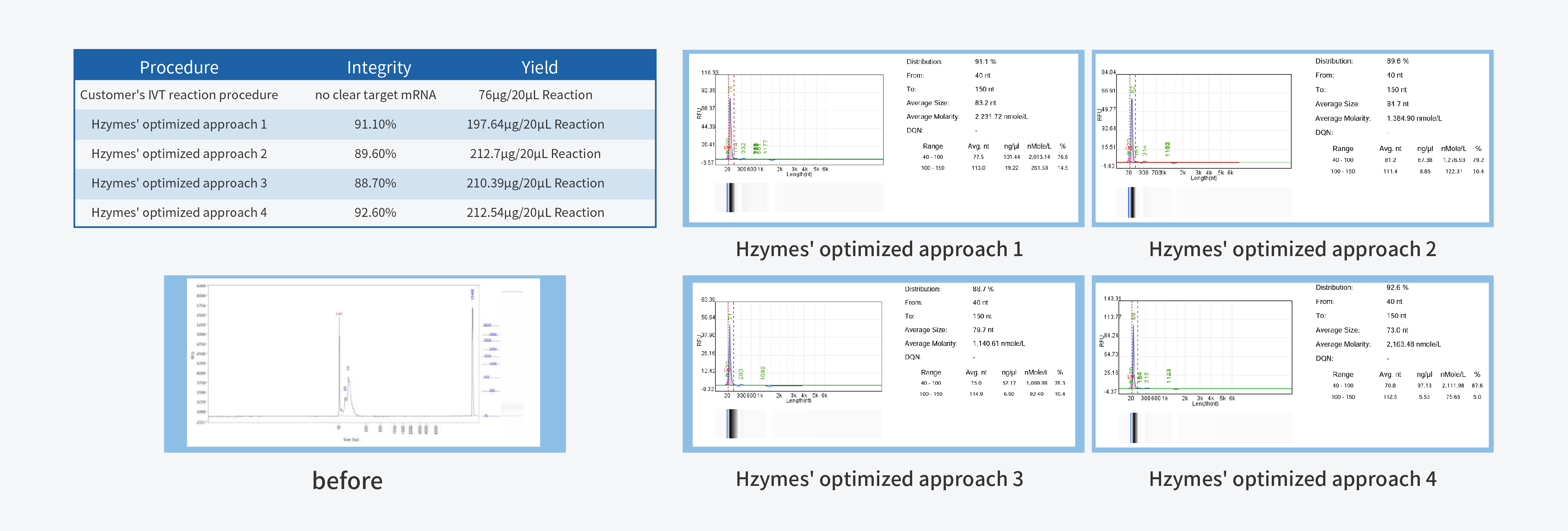
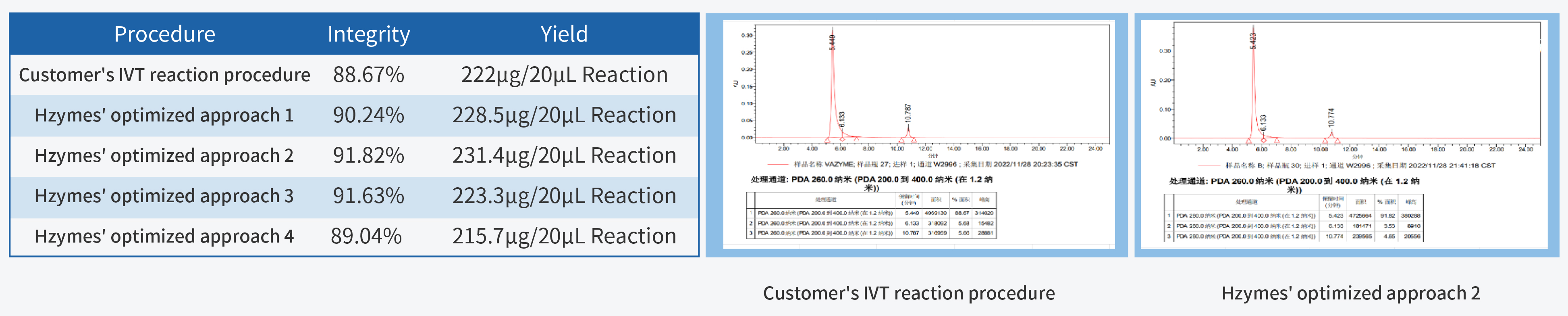
In the process of IVT (in vitro transcription) process development, achieving high yield and integrity are the primary fundamental quality indicators. High yield is closely linked to production costs, while high integrity forms the cornerstone of product quality.
T7 RNA Polymerase, a pivotal ingredient in IVT, plays a decisive role in mRNA quality. Hzymes, based on five core technology platforms involving enzyme gene resource exploration, directed evolution of enzyme, enzyme artificial intelligence design, large-scale enzyme manufacturing, and enzyme catalytic system application, offers a complete set of ingredients for mRNA synthesis.

RUN | Test | Load | UFDF pool filter |
1 | entrapment efficiency(%) | 97.66 | 94.84 |
| size(nm) | 67.37 | 71.51 | |
| PDI | 0.09 | 0.12 | |
2 | entrapment efficiency(%) | 95.69 | 95.00 |
| size(nm) | 66.90 | 73.94 | |
| PDI | 0.13 | 0.16 |




The foundation of the LNP encapsulation process is the design and development of the delivery system. A well-designed delivery system is necessary to prevent mRNA molecules from being degraded by RNase after entering the human body, effectively delivered to the target site, passed through the cell membrane, and released within the cell. Currently, Hzymes adopts the mainstream delivery system for lipid nanoparticles.
The key to the preparation of mRNA-LNP encapsulation is not only the formulation of lipid components, but also the control of the process, that is, how to control the contact and interaction between mRNA and lipid components to form stable, uniform, and high yield mRNA-LNP complexes. At present, Hzymes also adopts the current mainstream Microfluidics mixing technology. Due to the solubility of mRNA in the slightly acidic aqueous phase and the solubility of liposomes in ethanol, the mRNA solution and liposome solution are mixed by two opposing jets under high pressure. The strong turbulence causes each component to fully mix, while the ethanol phase is diluted and the pH of the solution changes. Liposomes precipitate to form lipid nanoparticles and form encapsulation complexes with mRNA.
After mRNA encapsulation, it is necessary to purify and remove unencapsulated/unloaded mRNA, free polymers or lipid materials, and adjust the final complex concentration, exchange the solvent buffer system, adjust pH value, etc. This step is usually achieved through tangential flow filtration, where the mRNA-LNP complex is intercepted, impurities and solvents are washed and filtered. Taking 4000nt mRNA as an example, it was diluted 20 times after encapsulation and perform tangential flow filtration using a hollow fiber column.
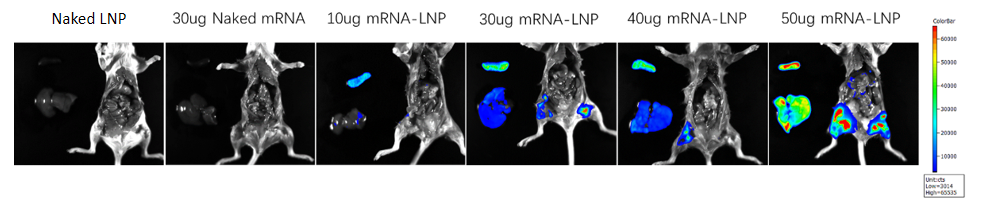
Delivering firefly luciferase mRNA-LNP into mice, the data showed that mRNA-LNP was successfully expressed in situ, targeting the liver and spleen.



For 4000nt mRNA, the purification process related data of Case is as follows:
Step | Cycle No. | Load | Elution | |||||||||
CE (%) | SEC_HPLC (H/M/L) (%) | Res. Protein (ug/mg) | Yield (%) | CE (%) | SEC_HPLC (H/M/L) (%) | Res. Protein (ug/mg) | dsRNA (ug/mg) | HCD (ug/mg) | Res.DNA template (ug/mg) | Endotoxin (EU/mg) | ||
TFF 1 | Cycle 1 | 76 | - | - | 100 | 78.8 | ND/97.3/2.8 | 8.5 | 0.69 | ND | 2.07E-15 | <9.4 |
AC | Cycle 1 | 71.5 | ND/97.9/2.1 | 6.3 | 92.1 | 82.5 | 3.7/95.1/1.2 | 0.2 | 0.60 | ND | 1.04E-15 | <17.5 |
Cycle 2 | 76.7 | ND/96.9/3.1 | 6.3 | 94.0 | 83.0 | 0.8/98.0/1.2 | 0.2 | 0.94 | ND | 9.36E-16 | <17.2 | |
TFF 2 | Cycle 1 | 82.8 | 3.6/95.4/0.9 | 0.2 | 99.6 | 82.5 | ND/99.6/0.4 | / | 0.68 | / | / | <6.8 |




Purification Process and Function of Hzymes mRNA Downstream Process
In general, the purification design of mRNA downstream process includes the following four Unit operation, which can be used to remove enzyme reactants, residual DNA and unwanted high molecular weight (HMW) substances.
· The first step is UF/DF filtration. Start from receiving IVT reaction products, then install UF/DF filters, flush with Nuclease free water, and balance buffer solution. Next, the product is concentrated to the target volume and exchanged into a buffer solution. Finally, the product is recovered by cleaning the UF/DF components. The purpose of this operation is to reduce impurities such as RNA polymerase, DNA template, NTPs, capping enzymes and reagents, inhibitors, and to exchange the buffer system.
· The second step is to use oligo dT resin to capture mRNA or use multimodal chromatography resin. The former uses oligo dT (polythymidine) ligand (18-25 bp Thymine deoxynucleotide) and polyadenylate (PolyA tail) at the 'end of mRNA 3' to form stable hybridization through hydrogen bond to capture mRNA. The purpose of this operation is to remove impurities lacking PloyA tail, such as free nucleotides, short chain transcripts, enzymes, etc. The latter is the mRNA flow through resin, which collects the flow through and rinse solution into a single fraction and proceeds to the next step. The purpose of this step is to remove the IVT reaction components contaminants.
· The third step is UF/DF filtration for concentration and buffer exchange. Replace the target product into the final formulation buffer and top wash the UF/DF components to recover the product.
· The final step is 0.2 µm sterilization and filtration. Then fill the closed drug substrate solution into a sterile container. It should be noted that any pipeline, chromatography or filter components that cannot be detected as free of nuclease should be kept with 0.5M NaOH for at least 1 hour under environmental conditions, and then washed with nuclease free water and appropriate buffer solution for the next Unit operation.
Finally, these purified mRNA drug substrates can be further encapsulated into LNP.
Purification process development and data display
In the field of mRNA, in addition to designing optimized mRNA sequences and developing efficient and safe delivery vectors, the development of simple, fast, large-scale, and cost-effective mRNA preparation processes is also one of the most important innovations in mRNA technology. In the production process of mRNA, purification after IVT is crucial for the safety and effectiveness of the final mRNA product, as the content of impurities can affect the translation efficiency of mRNA and alter immunogenicity. Therefore, it is necessary to remove impurities, including residual substrates, dsRNA, abnormal mRNA, DNA templates, protease residues, etc. In general, downstream purification methods include ultrafiltration, chromatography, precipitation, etc.













